
Difference: DPMatterStudies (1 vs. 7)
Revision 72018-09-14 - AlessioSarti
Dose Profiler matter studies pageRando Full Simulation studiesThe file chain is the following:
| |||||||||||
| Added: | |||||||||||
| > > |
To compute weights, on the selected tracks, run FRED.
/home_arpg/mfischetti/MeMs/MLEM/plugin
To run it
| ||||||||||
The weights definition + machineryHere we document how the weights are defined, where is the codeHow to run simulations and produce files to apply weights:To Run V2: -reference cilinder, necessary if the target does not start at 0cm (as reference cilinder3 does) -$FLUKA/flutil/rfluka -e fluka_FILTER_mbcil.exe -N0 -M5 filter_mbcil_1R & $FLUKA/flutil/rfluka -e fluka_FILTER_mbcil.exe -N0 -M5 filter_mbcil_2R & $FLUKA/flutil/rfluka -e fluka_FILTER_mbcil.exe -N0 -M5 filter_mbcil_3R & $FLUKA/flutil/rfluka -e fluka_FILTER_mbcil.exe -N0 -M5 filter_mbcil_4R & -mb brain+bone radius_ext = 10 cm centered in (0,0,10) $FLUKA/flutil/rfluka -e fluka_FILTER_mbfull.exe -N0 -M5 filter_mbfull_1R & $FLUKA/flutil/rfluka -e fluka_FILTER_mbfull.exe -N0 -M5 filter_mbfull_2R & $FLUKA/flutil/rfluka -e fluka_FILTER_mbfull.exe -N0 -M5 filter_mbfull_3R & $FLUKA/flutil/rfluka -e fluka_FILTER_mbfull.exe -N0 -M5 filter_mbfull_4R & cat filter_mbcil*00*dat > filter_mbcil_all_TXT.dat & cat filter_mbfull*00*dat > filter_mbfull_all_TXT.dat & ./Txt2Root -in filter_mbcil_all_TXT.dat -out filter_mbcil_all.root & Then in ../ApplyWeights: see ListApplyWeights _mbfull.sh (the first part) ->to have reference hZ distr Then: cd ../V5 ln -s ../V2/filter_mbfull_all_TXT.dat secondary_TXT.dat and run FLUKA on new ray*inp files (same full geo, different beam (10TeV muon+) and physics card) NB: check the source for reso! $FLUKA/flutil/rfluka -e fluka_RAY.exe -N0 -M1 ray_filter_mbfull_reso0 & $FLUKA/flutil/rfluka -e fluka_RAY.exe -N0 -M1 ray_filter_mbfull_reso1 & $FLUKA/flutil/rfluka -e fluka_RAY.exe -N0 -M1 ray_filter_mbfull_reso2 & $FLUKA/flutil/rfluka -e fluka_RAY.exe -N0 -M1 ray_filter_mbfull_reso3 & ./Txt2Root -in ray_filter_mbfull_reso0001_TXT.dat -out ray_filter_mbfull_reso0.root & ./Txt2Root -in ray_filter_mbfull_reso1001_TXT.dat -out ray_filter_mbfull_reso1.root & ./Txt2Root -in ray_filter_mbfull_reso2001_TXT.dat -out ray_filter_mbfull_reso2.root & ./Txt2Root -in ray_filter_mbfull_reso3001_TXT.dat -out ray_filter_mbfull_reso3.root & Then in ApplyWeights: see ListApplyWeights _mbfull.sh (second part) to produce the weighted Z distr - Compare the distributions with TestWeighs _mb.cc o quello che hai <3 The mental ball configurationsBeam: 12C ions, energy: 300 MeV /u, spatial distribution: gaussian with FWHM_x,y = 0.5 cm direction: positive z axis, starting point:(0,0,4) cm 1)*mbfull, REFERENCE distr: mbcil* Mental Ball: sphere with rmin = 9.6 cm, rmax = 10 cm, inner: BRAIN (ICRU), outer: BONE (ICRU) center:(0,0,15) cm - fluka region number of detectors (to be implemented in ApplyWeightsLib.h): {3,4,5,6,7,8} REFERENCE distr: mbcil = MB with same geo, material: water, cut with cilinder of r = 1.5 cm 2)*mbimp, REFERENCE distr: mbimp_cil* Mental Ball: sphere with rmin = 9.6 cm, rmax = 10 cm, inner: BRAIN (ICRU), outer: BONE (ICRU) center:(0,0,15) cm Implant: sphere, r = 1cm, material: AIR, center: (0.,5.,16.) - fluka region number of detectors (to be implemented in ApplyWeightsLib.h): {0,0,0,3,4,5} NB: the 60deg detectors are cut in order to have just a portion of ~ 20 x 20 cm2 ; no 90deg detectors in this configuration REFERENCE distr: mbimp_cil = MB with same geo, material: water, cut with cilinder of r = 1.5 cm (as in sim 1) ); detectors in same config of mbimp sim 3)*mbimp2, REFERENCE distr: mbimp_cil* Mental Ball: sphere with rmin = 9.6 cm, rmax = 10 cm, inner: BRAIN (ICRU), outer: BONE (ICRU) center:(0,0,15) cm Implant: sphere, r = 2cm, material: AIR, center: (0.,5.,16.) - fluka region number of detectors (to be implemented in ApplyWeightsLib.h): {0,0,0,3,4,5} NB: the 60deg detectors are cut in order to have just a portion of ~ 20 x 20 cm2 ; no 90deg detectors in this configuration REFERENCE distr: mbimp_cil = MB with same geo, material: water, cut with cilinder of r = 1.5 cm (as in sim 1) ); detectors in same config of mbimp sim 4)*mbimp3, REFERENCE distr: mbimp_cil* Mental Ball: sphere with rmin = 9.6 cm, rmax = 10 cm, inner: BRAIN (ICRU), outer: BONE (ICRU) center:(0,0,15) cm Implant: sphere, r = 1cm, material: ADIPOSE TISSUE, center: (0.,5.,16.) - fluka region number of detectors (to be implemented in ApplyWeightsLib.h): {0,0,0,3,4,5} NB: the 60deg detectors are cut in order to have just a portion of ~ 20 x 20 cm2 ; no 90deg detectors in this configuration REFERENCE distr: mbimp_cil = MB with same geo, material: water, cut with cilinder of r = 1.5 cm (as in sim 1) ); detectors in same config of mbimp sim Mental ball with inset on beam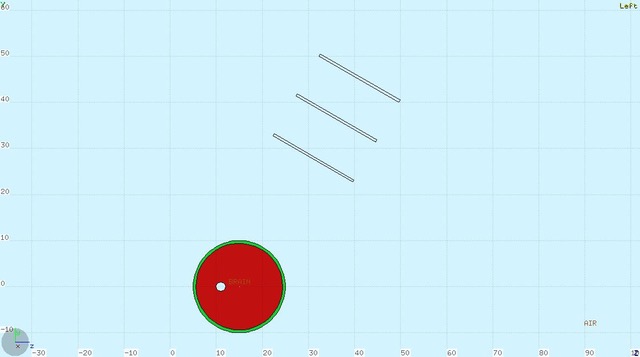
Comments
| |||||||||||
Revision 62018-09-14 - AlessioSarti
Dose Profiler matter studies pageThis page documents the matter studies. | |||||||||||
| Added: | |||||||||||
| > > | Rando Full Simulation studiesThe file chain is the following:
| ||||||||||
The weights definition + machineryHere we document how the weights are defined, where is the codeHow to run simulations and produce files to apply weights:To Run V2: -reference cilinder, necessary if the target does not start at 0cm (as reference cilinder3 does) -$FLUKA/flutil/rfluka -e fluka_FILTER_mbcil.exe -N0 -M5 filter_mbcil_1R & $FLUKA/flutil/rfluka -e fluka_FILTER_mbcil.exe -N0 -M5 filter_mbcil_2R & $FLUKA/flutil/rfluka -e fluka_FILTER_mbcil.exe -N0 -M5 filter_mbcil_3R & $FLUKA/flutil/rfluka -e fluka_FILTER_mbcil.exe -N0 -M5 filter_mbcil_4R & -mb brain+bone radius_ext = 10 cm centered in (0,0,10) $FLUKA/flutil/rfluka -e fluka_FILTER_mbfull.exe -N0 -M5 filter_mbfull_1R & $FLUKA/flutil/rfluka -e fluka_FILTER_mbfull.exe -N0 -M5 filter_mbfull_2R & $FLUKA/flutil/rfluka -e fluka_FILTER_mbfull.exe -N0 -M5 filter_mbfull_3R & $FLUKA/flutil/rfluka -e fluka_FILTER_mbfull.exe -N0 -M5 filter_mbfull_4R & cat filter_mbcil*00*dat > filter_mbcil_all_TXT.dat & cat filter_mbfull*00*dat > filter_mbfull_all_TXT.dat & ./Txt2Root -in filter_mbcil_all_TXT.dat -out filter_mbcil_all.root & Then in ../ApplyWeights: see ListApplyWeights _mbfull.sh (the first part) ->to have reference hZ distr Then: cd ../V5 ln -s ../V2/filter_mbfull_all_TXT.dat secondary_TXT.dat and run FLUKA on new ray*inp files (same full geo, different beam (10TeV muon+) and physics card) NB: check the source for reso! $FLUKA/flutil/rfluka -e fluka_RAY.exe -N0 -M1 ray_filter_mbfull_reso0 & $FLUKA/flutil/rfluka -e fluka_RAY.exe -N0 -M1 ray_filter_mbfull_reso1 & $FLUKA/flutil/rfluka -e fluka_RAY.exe -N0 -M1 ray_filter_mbfull_reso2 & $FLUKA/flutil/rfluka -e fluka_RAY.exe -N0 -M1 ray_filter_mbfull_reso3 & ./Txt2Root -in ray_filter_mbfull_reso0001_TXT.dat -out ray_filter_mbfull_reso0.root & ./Txt2Root -in ray_filter_mbfull_reso1001_TXT.dat -out ray_filter_mbfull_reso1.root & ./Txt2Root -in ray_filter_mbfull_reso2001_TXT.dat -out ray_filter_mbfull_reso2.root & ./Txt2Root -in ray_filter_mbfull_reso3001_TXT.dat -out ray_filter_mbfull_reso3.root & Then in ApplyWeights: see ListApplyWeights _mbfull.sh (second part) to produce the weighted Z distr - Compare the distributions with TestWeighs _mb.cc o quello che hai <3 The mental ball configurationsBeam: 12C ions, energy: 300 MeV /u, spatial distribution: gaussian with FWHM_x,y = 0.5 cm direction: positive z axis, starting point:(0,0,4) cm 1)*mbfull, REFERENCE distr: mbcil* Mental Ball: sphere with rmin = 9.6 cm, rmax = 10 cm, inner: BRAIN (ICRU), outer: BONE (ICRU) center:(0,0,15) cm - fluka region number of detectors (to be implemented in ApplyWeightsLib.h): {3,4,5,6,7,8} REFERENCE distr: mbcil = MB with same geo, material: water, cut with cilinder of r = 1.5 cm 2)*mbimp, REFERENCE distr: mbimp_cil* Mental Ball: sphere with rmin = 9.6 cm, rmax = 10 cm, inner: BRAIN (ICRU), outer: BONE (ICRU) center:(0,0,15) cm | |||||||||||
| Changed: | |||||||||||
| < < | Implant: sphere, r = 1cm, material: AIR, center: (0.,5.,16.) | ||||||||||
| > > | Implant: sphere, r = 1cm, material: AIR, center: (0.,5.,16.) | ||||||||||
| - fluka region number of detectors (to be implemented in ApplyWeightsLib.h): {0,0,0,3,4,5} NB: the 60deg detectors are cut in order to have just a portion of ~ 20 x 20 cm2 ; no 90deg detectors in this configuration REFERENCE distr: mbimp_cil = MB with same geo, material: water, cut with cilinder of r = 1.5 cm (as in sim 1) ); detectors in same config of mbimp sim 3)*mbimp2, REFERENCE distr: mbimp_cil* Mental Ball: sphere with rmin = 9.6 cm, rmax = 10 cm, inner: BRAIN (ICRU), outer: BONE (ICRU) center:(0,0,15) cm | |||||||||||
| Changed: | |||||||||||
| < < | Implant: sphere, r = 2cm, material: AIR, center: (0.,5.,16.) | ||||||||||
| > > | Implant: sphere, r = 2cm, material: AIR, center: (0.,5.,16.) | ||||||||||
| - fluka region number of detectors (to be implemented in ApplyWeightsLib.h): {0,0,0,3,4,5} NB: the 60deg detectors are cut in order to have just a portion of ~ 20 x 20 cm2 ; no 90deg detectors in this configuration REFERENCE distr: mbimp_cil = MB with same geo, material: water, cut with cilinder of r = 1.5 cm (as in sim 1) ); detectors in same config of mbimp sim 4)*mbimp3, REFERENCE distr: mbimp_cil* Mental Ball: sphere with rmin = 9.6 cm, rmax = 10 cm, inner: BRAIN (ICRU), outer: BONE (ICRU) center:(0,0,15) cm | |||||||||||
| Changed: | |||||||||||
| < < | Implant: sphere, r = 1cm, material: ADIPOSE TISSUE, center: (0.,5.,16.) | ||||||||||
| > > | Implant: sphere, r = 1cm, material: ADIPOSE TISSUE, center: (0.,5.,16.) | ||||||||||
| - fluka region number of detectors (to be implemented in ApplyWeightsLib.h): {0,0,0,3,4,5} NB: the 60deg detectors are cut in order to have just a portion of ~ 20 x 20 cm2 ; no 90deg detectors in this configuration REFERENCE distr: mbimp_cil = MB with same geo, material: water, cut with cilinder of r = 1.5 cm (as in sim 1) ); detectors in same config of mbimp sim Mental ball with inset on beam | |||||||||||
| Changed: | |||||||||||
| < < |  | ||||||||||
| > > |  | ||||||||||
Comments
| |||||||||||
Revision 52018-03-01 - AlessioSarti
Dose Profiler matter studies pageThe weights definition + machineryHere we document how the weights are defined, where is the codeHow to run simulations and produce files to apply weights:To Run V2: -reference cilinder, necessary if the target does not start at 0cm (as reference cilinder3 does) -$FLUKA/flutil/rfluka -e fluka_FILTER_mbcil.exe -N0 -M5 filter_mbcil_1R & $FLUKA/flutil/rfluka -e fluka_FILTER_mbcil.exe -N0 -M5 filter_mbcil_2R & $FLUKA/flutil/rfluka -e fluka_FILTER_mbcil.exe -N0 -M5 filter_mbcil_3R & $FLUKA/flutil/rfluka -e fluka_FILTER_mbcil.exe -N0 -M5 filter_mbcil_4R & -mb brain+bone radius_ext = 10 cm centered in (0,0,10) $FLUKA/flutil/rfluka -e fluka_FILTER_mbfull.exe -N0 -M5 filter_mbfull_1R & $FLUKA/flutil/rfluka -e fluka_FILTER_mbfull.exe -N0 -M5 filter_mbfull_2R & $FLUKA/flutil/rfluka -e fluka_FILTER_mbfull.exe -N0 -M5 filter_mbfull_3R & $FLUKA/flutil/rfluka -e fluka_FILTER_mbfull.exe -N0 -M5 filter_mbfull_4R & cat filter_mbcil*00*dat > filter_mbcil_all_TXT.dat & cat filter_mbfull*00*dat > filter_mbfull_all_TXT.dat & ./Txt2Root -in filter_mbcil_all_TXT.dat -out filter_mbcil_all.root & Then in ../ApplyWeights: | |||||||||||
| Changed: | |||||||||||
| < < | see ListApplyWeights _mbfull.sh (the first part) ->to have reference hZ distr | ||||||||||
| > > | see ListApplyWeights _mbfull.sh (the first part) ->to have reference hZ distr | ||||||||||
|
Then:
cd ../V5
ln -s ../V2/filter_mbfull_all_TXT.dat secondary_TXT.dat
and run FLUKA on new ray*inp files (same full geo, different beam (10TeV muon+) and physics card) NB: check the source for reso! $FLUKA/flutil/rfluka -e fluka_RAY.exe -N0 -M1 ray_filter_mbfull_reso0 & $FLUKA/flutil/rfluka -e fluka_RAY.exe -N0 -M1 ray_filter_mbfull_reso1 & $FLUKA/flutil/rfluka -e fluka_RAY.exe -N0 -M1 ray_filter_mbfull_reso2 & $FLUKA/flutil/rfluka -e fluka_RAY.exe -N0 -M1 ray_filter_mbfull_reso3 & ./Txt2Root -in ray_filter_mbfull_reso0001_TXT.dat -out ray_filter_mbfull_reso0.root & ./Txt2Root -in ray_filter_mbfull_reso1001_TXT.dat -out ray_filter_mbfull_reso1.root & ./Txt2Root -in ray_filter_mbfull_reso2001_TXT.dat -out ray_filter_mbfull_reso2.root & ./Txt2Root -in ray_filter_mbfull_reso3001_TXT.dat -out ray_filter_mbfull_reso3.root & Then in ApplyWeights: see ListApplyWeights _mbfull.sh (second part) to produce the weighted Z distr - Compare the distributions with TestWeighs _mb.cc o quello che hai <3 The mental ball configurationsBeam: 12C ions, energy: 300 MeV /u, spatial distribution: gaussian with FWHM_x,y = 0.5 cm direction: positive z axis, starting point:(0,0,4) cm 1)*mbfull, REFERENCE distr: mbcil* Mental Ball: sphere with rmin = 9.6 cm, rmax = 10 cm, inner: BRAIN (ICRU), outer: BONE (ICRU) center:(0,0,15) cm | |||||||||||
| Changed: | |||||||||||
| < < | - fluka region number of detectors (to be implemented in ApplyWeightsLib.h): {3,4,5,6,7,8} | ||||||||||
| > > | - fluka region number of detectors (to be implemented in ApplyWeightsLib.h): {3,4,5,6,7,8} | ||||||||||
| REFERENCE distr: mbcil = MB with same geo, material: water, cut with cilinder of r = 1.5 cm 2)*mbimp, REFERENCE distr: mbimp_cil* Mental Ball: sphere with rmin = 9.6 cm, rmax = 10 cm, inner: BRAIN (ICRU), outer: BONE (ICRU) center:(0,0,15) cm | |||||||||||
| Changed: | |||||||||||
| < < | Implant: sphere, r = 1cm, material: AIR, center: (0.,5.,16.) | ||||||||||
| > > | Implant: sphere, r = 1cm, material: AIR, center: (0.,5.,16.) | ||||||||||
| Changed: | |||||||||||
| < < | - fluka region number of detectors (to be implemented in ApplyWeightsLib.h): {0,0,0,3,4,5} | ||||||||||
| > > | - fluka region number of detectors (to be implemented in ApplyWeightsLib.h): {0,0,0,3,4,5} | ||||||||||
| Changed: | |||||||||||
| < < | NB: the 60deg detectors are cut in order to have just a portion of ~ 20 x 20 cm2 ; no 90deg detectors in this configuration | ||||||||||
| > > | NB: the 60deg detectors are cut in order to have just a portion of ~ 20 x 20 cm2 ; no 90deg detectors in this configuration | ||||||||||
| REFERENCE distr: mbimp_cil = MB with same geo, material: water, cut with cilinder of r = 1.5 cm (as in sim 1) ); detectors in same config of mbimp sim 3)*mbimp2, REFERENCE distr: mbimp_cil* Mental Ball: sphere with rmin = 9.6 cm, rmax = 10 cm, inner: BRAIN (ICRU), outer: BONE (ICRU) center:(0,0,15) cm | |||||||||||
| Changed: | |||||||||||
| < < | Implant: sphere, r = 2cm, material: AIR, center: (0.,5.,16.) | ||||||||||
| > > | Implant: sphere, r = 2cm, material: AIR, center: (0.,5.,16.) | ||||||||||
| Changed: | |||||||||||
| < < | - fluka region number of detectors (to be implemented in ApplyWeightsLib.h): {0,0,0,3,4,5} | ||||||||||
| > > | - fluka region number of detectors (to be implemented in ApplyWeightsLib.h): {0,0,0,3,4,5} | ||||||||||
| Changed: | |||||||||||
| < < | NB: the 60deg detectors are cut in order to have just a portion of ~ 20 x 20 cm2 ; no 90deg detectors in this configuration | ||||||||||
| > > | NB: the 60deg detectors are cut in order to have just a portion of ~ 20 x 20 cm2 ; no 90deg detectors in this configuration | ||||||||||
| REFERENCE distr: mbimp_cil = MB with same geo, material: water, cut with cilinder of r = 1.5 cm (as in sim 1) ); detectors in same config of mbimp sim 4)*mbimp3, REFERENCE distr: mbimp_cil* Mental Ball: sphere with rmin = 9.6 cm, rmax = 10 cm, inner: BRAIN (ICRU), outer: BONE (ICRU) center:(0,0,15) cm | |||||||||||
| Changed: | |||||||||||
| < < | Implant: sphere, r = 1cm, material: ADIPOSE TISSUE, center: (0.,5.,16.) | ||||||||||
| > > | Implant: sphere, r = 1cm, material: ADIPOSE TISSUE, center: (0.,5.,16.) | ||||||||||
| Changed: | |||||||||||
| < < | - fluka region number of detectors (to be implemented in ApplyWeightsLib.h): {0,0,0,3,4,5} | ||||||||||
| > > | - fluka region number of detectors (to be implemented in ApplyWeightsLib.h): {0,0,0,3,4,5} | ||||||||||
| Changed: | |||||||||||
| < < | NB: the 60deg detectors are cut in order to have just a portion of ~ 20 x 20 cm2 ; no 90deg detectors in this configuration | ||||||||||
| > > | NB: the 60deg detectors are cut in order to have just a portion of ~ 20 x 20 cm2 ; no 90deg detectors in this configuration | ||||||||||
| REFERENCE distr: mbimp_cil = MB with same geo, material: water, cut with cilinder of r = 1.5 cm (as in sim 1) ); detectors in same config of mbimp sim | |||||||||||
| Added: | |||||||||||
| > > | Mental ball with inset on beam
 | ||||||||||
Comments | |||||||||||
| Added: | |||||||||||
| > > |
| ||||||||||
Revision 42018-02-28 - IlariaMattei
Dose Profiler matter studies page | ||||||||
| Changed: | ||||||||
| < < | ||||||||
| > > | ||||||||
This page documents the matter studies.
The weights definition + machineryHere we document how the weights are defined, where is the codeHow to run simulations and produce files to apply weights: | ||||||||
| Changed: | ||||||||
| < < | To Run V2: | |||||||
| > > | To Run V2: | |||||||
| Changed: | ||||||||
| < < | -reference cilinder, but not necessary- $FLUKA/flutil/rfluka -e fluka_FILTER_mbcil.exe -N0 -M5 filter_mbcil_1R & $FLUKA/flutil/rfluka -e fluka_FILTER_mbcil.exe -N0 -M5 filter_mbcil_2R & $FLUKA/flutil/rfluka -e fluka_FILTER_mbcil.exe -N0 -M5 filter_mbcil_3R & $FLUKA/flutil/rfluka -e fluka_FILTER_mbcil.exe -N0 -M5 filter_mbcil_4R & | |||||||
| > > | -reference cilinder, necessary if the target does not start at 0cm (as reference cilinder3 does) - $FLUKA/flutil/rfluka -e fluka_FILTER_mbcil.exe -N0 -M5 filter_mbcil_1R & $FLUKA/flutil/rfluka -e fluka_FILTER_mbcil.exe -N0 -M5 filter_mbcil_2R & $FLUKA/flutil/rfluka -e fluka_FILTER_mbcil.exe -N0 -M5 filter_mbcil_3R & $FLUKA/flutil/rfluka -e fluka_FILTER_mbcil.exe -N0 -M5 filter_mbcil_4R & | |||||||
|
-mb brain+bone radius_ext = 10 cm centered in (0,0,10) $FLUKA/flutil/rfluka -e fluka_FILTER_mbfull.exe -N0 -M5 filter_mbfull_1R & $FLUKA/flutil/rfluka -e fluka_FILTER_mbfull.exe -N0 -M5 filter_mbfull_2R & $FLUKA/flutil/rfluka -e fluka_FILTER_mbfull.exe -N0 -M5 filter_mbfull_3R & $FLUKA/flutil/rfluka -e fluka_FILTER_mbfull.exe -N0 -M5 filter_mbfull_4R & cat filter_mbcil*00*dat > filter_mbcil_all_TXT.dat & cat filter_mbfull*00*dat > filter_mbfull_all_TXT.dat & ./Txt2Root -in filter_mbcil_all_TXT.dat -out filter_mbcil_all.root & Then in ../ApplyWeights: | ||||||||
| Changed: | ||||||||
| < < | see ListApplyWeights _mbfull.sh (first part)->to have reference distr (not necessary) | |||||||
| > > | see ListApplyWeights _mbfull.sh (the first part) ->to have reference hZ distr | |||||||
|
Then:
cd ../V5
ln -s ../V2/filter_mbfull_all_TXT.dat secondary_TXT.dat
and run FLUKA on new ray*inp files (same full geo, different beam (10TeV muon+) and physics card) NB: check the source for reso! $FLUKA/flutil/rfluka -e fluka_RAY.exe -N0 -M1 ray_filter_mbfull_reso0 & $FLUKA/flutil/rfluka -e fluka_RAY.exe -N0 -M1 ray_filter_mbfull_reso1 & $FLUKA/flutil/rfluka -e fluka_RAY.exe -N0 -M1 ray_filter_mbfull_reso2 & $FLUKA/flutil/rfluka -e fluka_RAY.exe -N0 -M1 ray_filter_mbfull_reso3 & ./Txt2Root -in ray_filter_mbfull_reso0001_TXT.dat -out ray_filter_mbfull_reso0.root & ./Txt2Root -in ray_filter_mbfull_reso1001_TXT.dat -out ray_filter_mbfull_reso1.root & ./Txt2Root -in ray_filter_mbfull_reso2001_TXT.dat -out ray_filter_mbfull_reso2.root & ./Txt2Root -in ray_filter_mbfull_reso3001_TXT.dat -out ray_filter_mbfull_reso3.root & Then in ApplyWeights: see ListApplyWeights _mbfull.sh (second part) | ||||||||
| Added: | ||||||||
| > > | to produce the weighted Z distr | |||||||
| - Compare the distributions with TestWeighs _mb.cc o quello che hai <3 | ||||||||
| Changed: | ||||||||
| < < | The mental ball configuration | |||||||
| > > | The mental ball configurations | |||||||
| Changed: | ||||||||
| < < | 1) Mental Ball: rmin = 9.6 cm, rmax = 10 cm, inner: BRAIN (ICRU), outer: BONE (ICRU) | |||||||
| > > | Beam: 12C ions, energy: 300 MeV /u, spatial distribution: gaussian with FWHM_x,y = 0.5 cm | |||||||
| Changed: | ||||||||
| < < | center:(0,0,10) cm | |||||||
| > > | direction: positive z axis, starting point:(0,0,4) cm | |||||||
| Changed: | ||||||||
| < < | Beam: 12C ions, energy: 300 MeV /u, spatial distribution: gaussian with FWHM_x,y = 0.5 cm | |||||||
| > > | 1)*mbfull, REFERENCE distr: mbcil* | |||||||
| Changed: | ||||||||
| < < | direction: positive z axis, starting point:(0,0,-1) cm | |||||||
| > > | Mental Ball: sphere with rmin = 9.6 cm, rmax = 10 cm, inner: BRAIN (ICRU), outer: BONE (ICRU) | |||||||
| Changed: | ||||||||
| < < | -- | |||||||
| > > | center:(0,0,15) cm | |||||||
| Changed: | ||||||||
| < < | Comments | |||||||
| > > | - fluka region number of detectors (to be implemented in ApplyWeightsLib.h): {3,4,5,6,7,8} | |||||||
| Changed: | ||||||||
| < < | ||||||||
| > > | REFERENCE distr: mbcil = MB with same geo, material: water, cut with cilinder of r = 1.5 cm | |||||||
| Added: | ||||||||
| > > | 2)*mbimp, REFERENCE distr: mbimp_cil*
Mental Ball: sphere with rmin = 9.6 cm, rmax = 10 cm, inner: BRAIN (ICRU), outer: BONE (ICRU)
center:(0,0,15) cm
Implant: sphere, r = 1cm, material: AIR, center: (0.,5.,16.)
- fluka region number of detectors (to be implemented in ApplyWeightsLib.h): {0,0,0,3,4,5}
NB: the 60deg detectors are cut in order to have just a portion of ~ 20 x 20 cm2 ; no 90deg detectors in this configuration
REFERENCE distr: mbimp_cil = MB with same geo, material: water, cut with cilinder of r = 1.5 cm (as in sim 1) ); detectors in same config of mbimp sim
3)*mbimp2, REFERENCE distr: mbimp_cil*
Mental Ball: sphere with rmin = 9.6 cm, rmax = 10 cm, inner: BRAIN (ICRU), outer: BONE (ICRU)
center:(0,0,15) cm
Implant: sphere, r = 2cm, material: AIR, center: (0.,5.,16.)
- fluka region number of detectors (to be implemented in ApplyWeightsLib.h): {0,0,0,3,4,5}
NB: the 60deg detectors are cut in order to have just a portion of ~ 20 x 20 cm2 ; no 90deg detectors in this configuration
REFERENCE distr: mbimp_cil = MB with same geo, material: water, cut with cilinder of r = 1.5 cm (as in sim 1) ); detectors in same config of mbimp sim
4)*mbimp3, REFERENCE distr: mbimp_cil*
Mental Ball: sphere with rmin = 9.6 cm, rmax = 10 cm, inner: BRAIN (ICRU), outer: BONE (ICRU)
center:(0,0,15) cm
Implant: sphere, r = 1cm, material: ADIPOSE TISSUE, center: (0.,5.,16.)
- fluka region number of detectors (to be implemented in ApplyWeightsLib.h): {0,0,0,3,4,5}
NB: the 60deg detectors are cut in order to have just a portion of ~ 20 x 20 cm2 ; no 90deg detectors in this configuration
REFERENCE distr: mbimp_cil = MB with same geo, material: water, cut with cilinder of r = 1.5 cm (as in sim 1) ); detectors in same config of mbimp sim
Comments | |||||||
Revision 32018-02-23 - IlariaMattei
Dose Profiler matter studies page | ||||||||
| Changed: | ||||||||
| < < | ||||||||
| > > | ||||||||
This page documents the matter studies.
The weights definition + machineryHere we document how the weights are defined, where is the code | ||||||||
| Added: | ||||||||
| > > | How to run simulations and produce files to apply weights:To Run V2: -reference cilinder, but not necessary-$FLUKA/flutil/rfluka -e fluka_FILTER_mbcil.exe -N0 -M5 filter_mbcil_1R & $FLUKA/flutil/rfluka -e fluka_FILTER_mbcil.exe -N0 -M5 filter_mbcil_2R & $FLUKA/flutil/rfluka -e fluka_FILTER_mbcil.exe -N0 -M5 filter_mbcil_3R & $FLUKA/flutil/rfluka -e fluka_FILTER_mbcil.exe -N0 -M5 filter_mbcil_4R & -mb brain+bone radius_ext = 10 cm centered in (0,0,10) $FLUKA/flutil/rfluka -e fluka_FILTER_mbfull.exe -N0 -M5 filter_mbfull_1R & $FLUKA/flutil/rfluka -e fluka_FILTER_mbfull.exe -N0 -M5 filter_mbfull_2R & $FLUKA/flutil/rfluka -e fluka_FILTER_mbfull.exe -N0 -M5 filter_mbfull_3R & $FLUKA/flutil/rfluka -e fluka_FILTER_mbfull.exe -N0 -M5 filter_mbfull_4R & cat filter_mbcil*00*dat > filter_mbcil_all_TXT.dat & cat filter_mbfull*00*dat > filter_mbfull_all_TXT.dat & ./Txt2Root -in filter_mbcil_all_TXT.dat -out filter_mbcil_all.root & Then in ../ApplyWeights: see ListApplyWeights _mbfull.sh (first part)->to have reference distr (not necessary) Then: cd ../V5 ln -s ../V2/filter_mbfull_all_TXT.dat secondary_TXT.dat and run FLUKA on new ray*inp files (same full geo, different beam (10TeV muon+) and physics card) NB: check the source for reso! $FLUKA/flutil/rfluka -e fluka_RAY.exe -N0 -M1 ray_filter_mbfull_reso0 & $FLUKA/flutil/rfluka -e fluka_RAY.exe -N0 -M1 ray_filter_mbfull_reso1 & $FLUKA/flutil/rfluka -e fluka_RAY.exe -N0 -M1 ray_filter_mbfull_reso2 & $FLUKA/flutil/rfluka -e fluka_RAY.exe -N0 -M1 ray_filter_mbfull_reso3 & ./Txt2Root -in ray_filter_mbfull_reso0001_TXT.dat -out ray_filter_mbfull_reso0.root & ./Txt2Root -in ray_filter_mbfull_reso1001_TXT.dat -out ray_filter_mbfull_reso1.root & ./Txt2Root -in ray_filter_mbfull_reso2001_TXT.dat -out ray_filter_mbfull_reso2.root & ./Txt2Root -in ray_filter_mbfull_reso3001_TXT.dat -out ray_filter_mbfull_reso3.root & Then in ApplyWeights: see ListApplyWeights _mbfull.sh (second part) - Compare the distributions with TestWeighs _mb.cc o quello che hai <3 | |||||||
The mental ball configuration1) Mental Ball: rmin = 9.6 cm, rmax = 10 cm, inner: BRAIN (ICRU), outer: BONE (ICRU) | ||||||||
| Changed: | ||||||||
| < < | center:(0,0,10) cm | |||||||
| > > | center:(0,0,10) cm | |||||||
| Changed: | ||||||||
| < < | Beam: 12C ions, energy: 300 MeV/u, spatial distribution: gaussian with FWHM_x,y = 0.5 cm | |||||||
| > > | Beam: 12C ions, energy: 300 MeV /u, spatial distribution: gaussian with FWHM_x,y = 0.5 cm | |||||||
| Changed: | ||||||||
| < < | direction: positive z axis, starting point:(0,0,-1) cm | |||||||
| > > | direction: positive z axis, starting point:(0,0,-1) cm | |||||||
| Changed: | ||||||||
| < < | -- | |||||||
| > > | -- | |||||||
Comments | ||||||||
| Changed: | ||||||||
| < < | ||||||||
| > > | ||||||||
| Added: | ||||||||
| > > | ||||||||
Revision 22018-02-23 - IlariaMattei
Dose Profiler matter studies pageThe weights definition + machineryHere we document how the weights are defined, where is the codeThe mental ball configuration | ||||||||
| Changed: | ||||||||
| < < | What samples are available, what statistics | |||||||
| > > | 1) Mental Ball: rmin = 9.6 cm, rmax = 10 cm, inner: BRAIN (ICRU), outer: BONE (ICRU) | |||||||
| Changed: | ||||||||
| < < | -- | |||||||
| > > | center:(0,0,10) cm | |||||||
| Added: | ||||||||
| > > | Beam: 12C ions, energy: 300 MeV/u, spatial distribution: gaussian with FWHM_x,y = 0.5 cm
direction: positive z axis, starting point:(0,0,-1) cm
-- | |||||||
Comments | ||||||||
| Deleted: | ||||||||
| < < | ||||||||
Revision 12018-02-22 - AlessioSarti
Dose Profiler matter studies pageThe weights definition + machineryHere we document how the weights are defined, where is the codeThe mental ball configurationWhat samples are available, what statistics --Comments |
View topic | History: r7 < r6 < r5 < r4 | More topic actions...
Ideas, requests, problems regarding TWiki? Send feedback